Predicting Gene Expression with AI-powered Enformer and Machine Learning
Enformer leverages AI and Transformers to decode non-coding DNA, predicting gene expression with high accuracy. By capturing long-range interactions, Enformer reveals genetic variant impacts, advancing genomics and personalized medicine.

Introduction
Did you know that less than 2% of DNA in our whole genome codes for proteins, while the other 98% hide secrets in the non-coding DNA?
This non-coding DNA plays a crucial role in deciding when and where those protein-making genes should play their major role.
Think of genes as a set of instructions that build and run our bodies.
But just like a manual, these instructions need to be interpreted correctly.
That’s where gene expression comes in—it decides when, where, and how much of a gene's message should be used to make proteins.
This complex process is guided by hidden elements within our DNA, and understanding it can be challenging.
Understanding gene instructions isn't always easy. That's where annotation plays a crucial role.

Annotation helps scientists identify the parts of the gene sequence that control gene expression, telling us when and where certain genes should be active.
Knowing gene expression helps us understand how our bodies work and why we are unique.
This understanding reveals how our bodies function and highlights what makes each of us unique. Scientists can predict diseases early by studying gene expression patterns and developing strategies to keep us healthier.
In this blog, we will understand Enformer architecture made by Google Deepmind for Gene Expression.
Moreover, we will learn how Enformer helps discover why a certain genetic variant, rs11644125, might affect our immune system.
We will also explore Enformer's real-life applications and how it helps scientists predict changes in our DNA, called genetic variants, that might impact our health.
Table of Contents
- Introduction
- The Challenge of Non-Coding DNA
- Enformer Architecture: A Transformer Approach
- Enformer Use for Understanding Disease-Associated Variants
- Understanding the Predictive Ability
- Future Directions
- Conclusion
- Frequently Asked Questions
The Challenge of Non-Coding DNA

The remaining 98 non-coding DNA is often overlooked but incredibly important.
This non-coding DNA plays a pivotal role in our body's functioning.
This complex regulation is influenced by enhancers and other regulatory elements, which poses a challenge for traditional models.
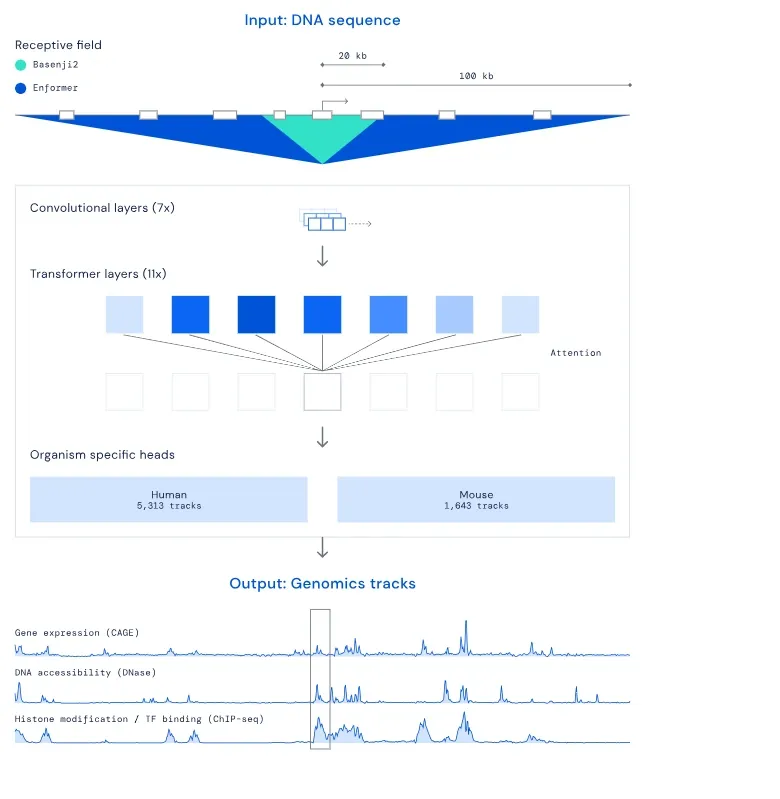
Enformer addresses this challenge by using Transformers, which is commonly used in natural language processing, to process and understand vast DNA sequences of up to 200,000 base pairs.
Enformer Architecture: A Transformer Approach
Enformer's departure from traditional convolutional neural networks (CNNs) in favor of Transformers is a strategic move that significantly enhances its ability to understand and predict gene expression patterns.
Let's see how Enformer achieves this and why it outperforms previous models.
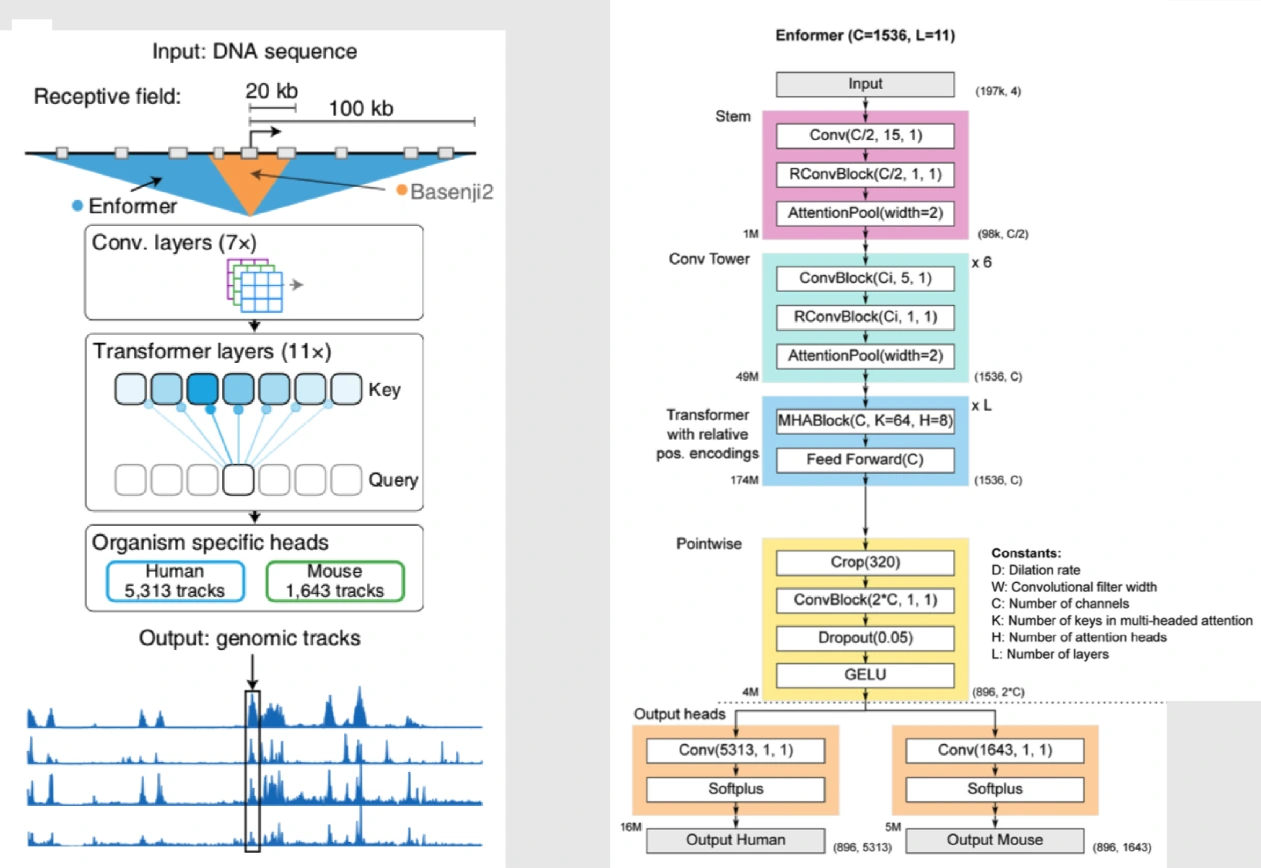
1. Transformer Architecture
The key innovation in Enformer lies in its adoption of Transformer which is a type of neural network architecture initially popularized in natural language processing tasks.
Transformers use self-attention mechanisms allowing them to focus on different parts of the input sequence when making predictions.
This is particularly beneficial in genomics where understanding long-range interactions within DNA sequences is crucial.
2. Capturing Long-Range Interactions
Traditional CNNs used commonly in gene expression prediction models, have limitations in capturing long-range dependencies within DNA sequences.
This is especially problematic when dealing with regulatory elements located far from the target gene.
Enformer addresses this limitation by leveraging the self-attention mechanisms inherent in Transformers, allowing it to consider interactions at much greater distances.
3. Expanded Receptive Field
Enformer's unique strength lies in its expanded receptive field.
This term refers to the range of input data that influences the predictions made by the model.
In the case of Enformer, its ability to consider interactions at distances more than five times greater than previous methods gives it a substantial advantage in modeling the complexities of non-coding regions.
4. Decoding Non-Coding Regions
The non-coding regions of the genome which make up the majority of DNA have been challenging for traditional models.
Enformer's adoption of Transformers enables it to effectively decode these non-coding regions by capturing the relationships and regulatory elements that influence gene expression.
The self-attention mechanisms allow Enformer to attend to relevant portions of the DNA sequence even when they are located far from the target gene.
5. Enhanced Predictive Accuracy
Enformer achieves unprecedented predictive accuracy with its expanded receptive field and the ability to capture long-range interactions.
It excels in predicting gene expression levels by considering a much broader context of the genome.
This accuracy is crucial in understanding gene regulation, especially when dealing with complex mechanisms influenced by distant enhancers and other regulatory elements.
Enformer use for understanding Disease-Associated Variants
Enformer is the advanced gene prediction model showcasing its incredible abilities in understanding a specific genetic variant linked to lower levels of certain white blood cells.
Scientists discovered a specific genetic variant known as rs11644125, which was associated with reduced levels of particular white blood cells in the body.
Understanding the impact of this variant on our genes could provide valuable insights into the mechanisms behind changes in white blood cell counts which is an essential aspect of our immune system.
Enformer's Work-flow
1. Identification of the Variant
Enformer first identified the genetic variant, rs11644125, known for its connection to lower white blood cell levels.
2. Systematic Mutations
Enformer systematically made changes to the positions surrounding the identified variant.
3. Predicting Gene Expression
Enformer forecasted how these changes in the DNA sequence would impact the expression of a specific gene called NLRC5.
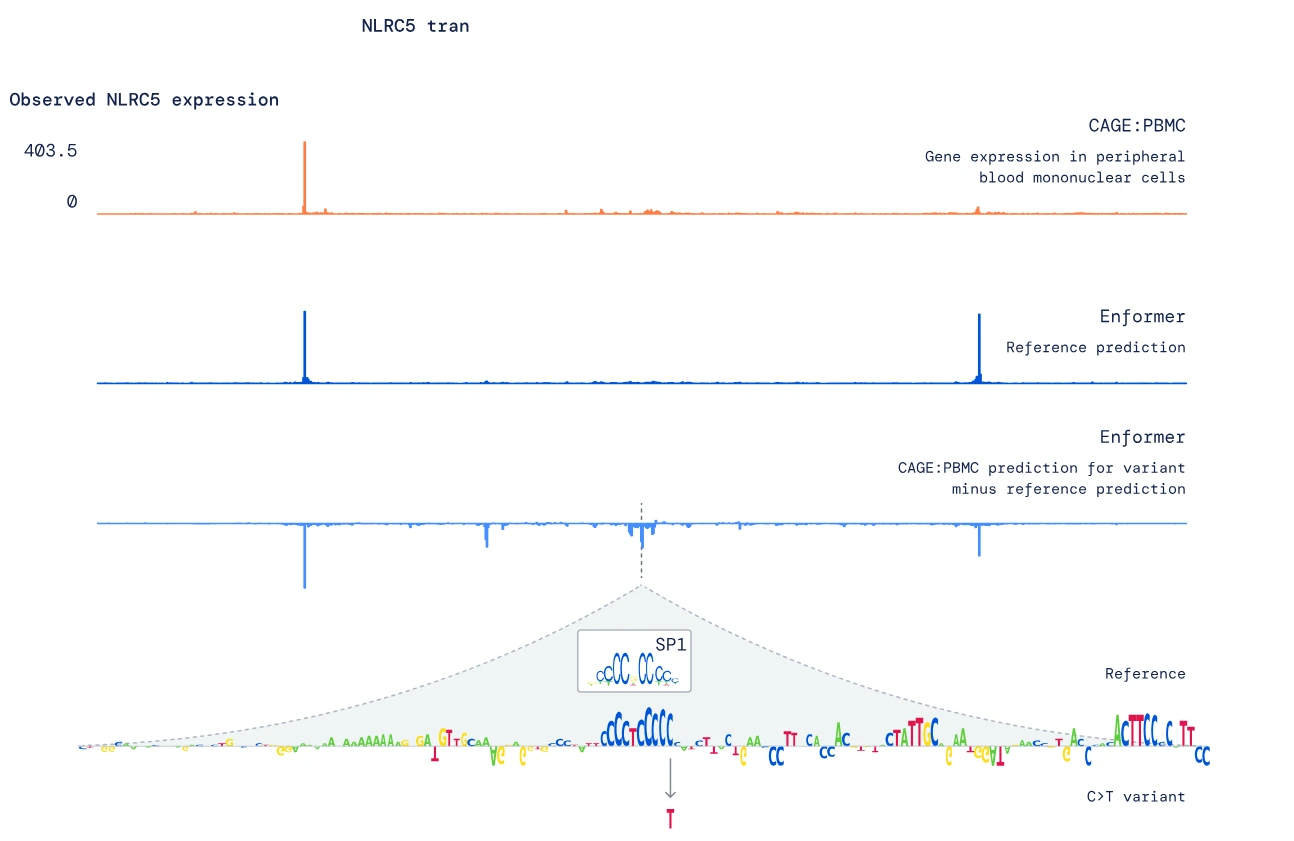
4. Insight into NLRC5 Gene
Enformer's analysis revealed that the rs11644125 variant had a notable effect on the NLRC5 gene's expression.
This gene is known to play a role in immune responses, particularly in the production of certain white blood cells.
5. Exploring the Mechanism
The findings suggested that the genetic variant influenced how the NLRC5 gene was expressed, ultimately leading to lower levels of specific white blood cells.
It's like Enformer acted as a detective, uncovering a potential biological mechanism behind changes in immune cell counts.
The Power of Enformer
Enformer's strength lies in its ability to decipher the impact of genetic variations on gene expression.
It was a helpful tool to figure out how the rs11644125 variant influences the NLRC5 gene and, as a result, our immune system.
This case study demonstrates how Enformer's ability to predict things is helping us learn more about differences in our genes.
This could lead to new discoveries in personalized medicine and specific treatments for health issues related to our immune system.
Application in Variant Analysis
One of the main applications of Enformer is predicting how changes to DNA letters, known as genetic variants, impact gene expression.
Enformer outperforms previous models in accuracy, particularly when analyzing natural and synthetic variants affecting crucial regulatory sequences.
This capability is crucial for interpreting disease-associated variants obtained through genome-wide association studies, as many disease-linked variants are located in the non-coding regions.
Understanding the Predictive Ability
Enformer is trained to predict functional genomic data, including gene expression, from 200,000 base pairs of input DNA.
Transformer modules within Enformer use attention mechanisms to process the entire sequence, effectively considering much longer input sequences compared to earlier models.
To understand how Enformer arrives at its predictions, contribution scores are used to highlight influential parts of the input sequence.
Notably, Enformer accurately identifies enhancers located more than 50,000 base pairs away from the gene, providing valuable insights into gene regulation.
Future Directions
The effort to understand the entire human genetic code isn't done yet, even though Enformer has significantly improved our understanding of the details of genomic sequences.
The application of AI and ML to genomics has the potential to advance our understanding of illnesses, uncover genomic patterns, and provide mechanistic theories.
Collaborations with scientists and institutions keen to use computational models for genomic exploration are essential as we explore further into the vast domain of the human genome.
Conclusion
Enformer's use of Transformers powered by Artificial Intelligence in predicting gene expression marks a change in the perspective of genetic research.
By overcoming the challenges posed by non-coding DNA, Enformer opens new possibilities for understanding the intricate language of the genome.
As AI and ML continue to play important roles in decoding genetic complexities, the path to personalized medicine and a precise understanding of disease mechanisms becomes clearer.
The teamwork of technology and genetics is opening the door to a future where we might finally solve the mysteries of the human genome.
FAQ
1. Can deep learning improve gene expression prediction accuracy?
Yes, deep learning, particularly through advanced models like Enformer, significantly enhances gene expression prediction accuracy.
Traditional models, often reliant on convolutional neural networks (CNNs), face challenges in capturing long-range interactions within DNA sequences, especially in non-coding regions.
Enformer's strategic use of Transformers, with self-attention mechanisms, allows it to consider interactions at much greater distances, leading to an expanded receptive field.
This breakthrough enables Enformer to achieve unprecedented predictive accuracy by considering a broader context of the genome.
The ability to capture long-range dependencies and decode non-coding regions contributes to its success in understanding the complexities of gene regulation, showcasing the power of deep learning in genomics.
2. Can machine learning predict gene expression?
Yes, machine learning, particularly advanced models like Enformer, can predict gene expression.
Enformer employs a Transformer-based architecture with self-attention mechanisms, enabling it to analyze and understand vast DNA sequences.
Its ability to capture long-range interactions and consider non-coding regions, where traditional models often struggle, results in great predictive accuracy.
By leveraging these machine learning techniques, Enformer can forecast how changes in DNA sequences influence gene expression, contributing to a deeper understanding of the complexities of genomic regulation.
3. How does enformer predict gene expression?
Enformer predicts gene expression by utilizing a Transformer-based architecture with self-attention mechanisms.
This innovative approach allows Enformer to process and understand extensive DNA sequences, considering long-range interactions crucial in genomics.
The self-attention mechanisms enable the model to focus on different parts of the input sequence when making predictions, capturing intricate relationships within the DNA.
Enformer can analyze the impact of genetic variations on gene expression, offering valuable insights into the complexities of genomic regulation.
Simplify Your Data Annotation Workflow With Proven Strategies
.png)