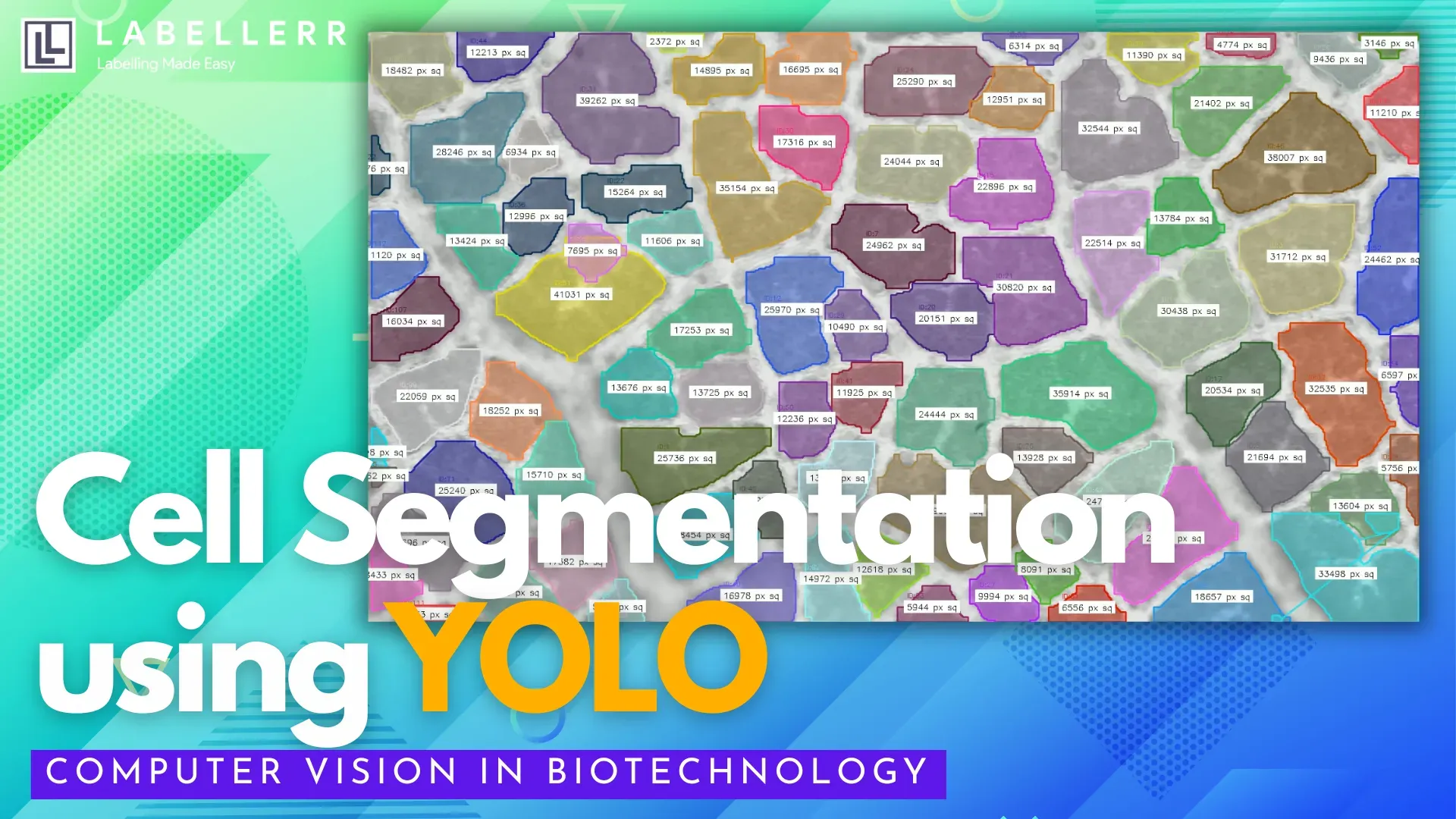
Building a Cell Segmentation System using Labellerr and YOLO
Computer vision brings speed and accuracy to cell segmentation, processing dense microscopy images rapidly. With Labellerr’s SAM-powered annotation and collaboration tools, researchers can build scalable, reliable AI models for biomedical imaging.
Picture a time-pressed scientist late at night, hunched over a microscope, staring at a field teeming with thousands of overlapping cells.
Every experiment means painstakingly counting, marking, and analyzing cells by hand, a process that is not only tedious but highly inconsistent.
In many labs, manual cell counting is still common, but it's plagued by errors, fatigue, and subjectivity.
Results can vary wildly between technicians, with small mistakes causing major setbacks in research.
This is a universal frustration for anyone working in biomedical imaging.
The Solution
Computer vision has emerged as a transformative ally, automating what was once slow, error-prone manual labor.
Leveraging deep learning, it can instantly interpret complex microscopy images and perform cell segmentation, the task of identifying and outlining unique cells within dense images.
But what exactly is cell segmentation? It’s a technique where AI models analyze pixel data to draw boundaries around every cell, even when cells overlap or blend into noisy backgrounds. This enables researchers to:
- Objectively measure cell morphology (size, shape, structure)
- Accurately count large numbers of cells
- Analyze spatial distribution within tissues
- Track dynamic behaviors in live-cell experiments
These capabilities are crucial for high-throughput studies on disease progression, drug efficacy, and tissue engineering, all of which demand reproducible and scalable data.
Speed and Scalability
Computer vision (CV) has dramatically improved the speed and scalability of cell segmentation in biomedical research.
Platforms like Ultralytics and Biodock use advanced deep learning models such as YOLOv8 to segment thousands of cellular images in minutes, compared to hours with manual or classical approaches.
For example, YOLO-based methods can process electron microscopy images up to 43 times faster than traditional models like U-Net, while cloud-based tools allow researchers to analyze experiments up to 3000 times faster than on local machines.
This acceleration makes large-scale analysis reliable and reproducible, enabling researchers to quickly unlock insights that drive scientific progress.
How Labellerr Helps Biotechnology?
Labellerr plays a pivotal role in accelerating and improving model training for cell segmentation by providing robust, high-quality data annotation services.
Using advanced algorithms and innovative tools like the Segment Anything Model (SAM), Labellerr allows researchers to efficiently and accurately annotate thousands of microscopic images, outlining individual cells, handling complex overlaps, and ensuring precise boundary identification.
Labellerr’s collaborative platform also streamlines teamwork, enabling multiple users to contribute to large projects and quickly build comprehensive datasets for machine learning in biomedical imaging and research.
By supporting automated import/export and offering cost-effective solutions, Labellerr ensures that organizations can scale up annotation efforts without compromising quality or research budgets, accelerating scientific discoveries in cell segmentation and medical AI.
Create Cell Segmentation Model using Labellerr
This code structure, based on the workflow from our research, shows how you can take the exported data from Labellerr and train your own YOLOv8 segmentation model.
import torch
from ultralytics import YOLO
import cv2
import numpy as np
import random
# --- 1. Data Preparation ---
# (This step is assumed to be complete after exporting from Labellerr)
# You would first run a script to convert your exported COCO JSON
# into the YOLO segmentation format. This creates a 'data.yaml' file
# and 'train/images', 'train/labels', 'valid/images', 'valid/labels' folders.
# --- 2. GPU Check & Setup ---
# Clear VRAM if you are using a GPU for training
if torch.cuda.is_available():
torch.cuda.empty_cache()
print("CUDA (GPU) is available. VRAM cleared.")
else:
print("CUDA is not available. Training will run on CPU.")
# --- 3. Model Training ---
# Load a pre-trained YOLOv8 segmentation model
model = YOLO('yolov8n-seg.pt')
# Train the model on your custom dataset
print("Starting model training...")
results = model.train(
data='path/to/your/dataset/data.yaml',
epochs=500,
imgsz=640,
batch=-1, # Automatically determines batch size
device=0, # Use GPU (device=0) if available
name='yolo_cell_segmentation_model' # Name for the results folder
)
print("Training complete. Model saved to 'runs/segment/...'")
# --- 4. Inference and Analysis Class ---
# Load your newly trained model
trained_model_path = 'runs/segment/train/weights/best.pt'
inference_model = YOLO(trained_model_path)
class CellSegmenter:
def __init__(self, model, conf_threshold=0.5, iou_threshold=0.3):
self.model = model
self.conf = conf_threshold
self.iou = iou_threshold
self.colors = {} # To store a random color for each cell ID
def _get_color(self, track_id):
# Assign a consistent random color to each tracked cell
if track_id not in self.colors:
self.colors[track_id] = (random.randint(0, 255), random.randint(0, 255), random.randint(0, 255))
return self.colors[track_id]
def _calculate_area(self, mask_contour):
# Calculate the area of the segmented polygon
return cv2.contourArea(mask_contour)
def process_frame(self, frame):
# Run tracking and segmentation on the frame
results = self.model.track(
frame,
persist=True,
conf=self.conf,
iou=self.iou
)
annotated_frame = frame.copy()
overlay = frame.copy()
if results[0].masks:
for r in results:
masks = r.masks.xy # Mask polygons
boxes = r.boxes # Bounding boxes with tracking IDs
for mask, box in zip(masks, boxes):
if box.id is not None:
track_id = int(box.id.item())
color = self._get_color(track_id)
# Draw the segmentation mask
mask_contour = np.array(mask, dtype=np.int32)
cv2.fillPoly(overlay, [mask_contour], color)
# Calculate area
area = self._calculate_area(mask_contour)
# Get position for text
x1, y1, x2, y2 = map(int, box.xyxy[0])
text_pos = (x1, y1 - 10)
# Draw label with area
label = f'Cell ID: {track_id} Area: {int(area)}'
cv2.putText(annotated_frame, label, text_pos,
cv2.FONT_HERSHEY_SIMPLEX, 0.5, color, 2)
cv2.rectangle(annotated_frame, (x1,y1), (x2,y2), color, 2)
# Create a transparent overlay
cv2.addWeighted(overlay, 0.4, annotated_frame, 0.6, 0, annotated_frame)
return annotated_frame
def process_video(self, video_path, output_path):
cap = cv2.VideoCapture(video_path)
# Get video properties for writer
w = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
h = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
fps = int(cap.get(cv2.CAP_PROP_FPS))
fourcc = cv2.VideoWriter_fourcc(*'mp4v')
out = cv2.VideoWriter(output_path, fourcc, fps, (w, h))
print("Processing video...")
while cap.isOpened():
ret, frame = cap.read()
if not ret:
break
processed_frame = self.process_frame(frame)
out.write(processed_frame)
cap.release()
out.release()
cv2.destroyAllWindows()
print(f"Video processing complete. Saved to {output_path}")
# --- 5. Example Usage ---
segmenter = CellSegmenter(model=inference_model)
segmenter.process_video(
video_path='path/to/your/sample_video.mp4',
output_path='output/cell_analysis_video.mp4'
)
Conclusion
The power of computer vision to revolutionize biotechnology is clear. It moves scientific analysis from a slow, manual process to a fast, data-driven one. However, this advanced AI is not built on complex code alone.
The true foundation of a successful, specialized model is high-quality, accurately annotated data. The code is the vehicle, but the data is the fuel. The importance of the data annotation step cannot be overstated, it is the single most critical factor in determining whether a model will be a powerful tool for discovery or a failed experiment.
FAQs
Why is manual cell segmentation unreliable in high-throughput experiments?
Manual methods suffer from inconsistency, fatigue, and subjectivity, limiting accuracy for large-scale biomedical imaging studies.
How does computer vision enhance scalability in cell segmentation?
Deep learning models segment thousands of dense images rapidly, enabling fast and reproducible analysis of large datasets.
What role does Labellerr play in training segmentation models?
Labellerr supports high-quality data creation with SAM-assisted annotation tools, collaboration features, and efficient dataset management.
Simplify Your Data Annotation Workflow With Proven Strategies
.png)